This first tutorial will guide you through the process of setting up a new application using the DiFfRG. This first program will simply solve a hydrodynamic equation unrelated to fRG, but introduce the basic structure of a typical DiFfRG application.
You can find the full code as described here also in Tutorials/tut1.
File structure
We will start by creating a new directory in the Applications directory just below the DiFfRG top directory. This directory will contain all the files necessary for the simulation.
- The
CMakeLists.txtfile is used to tell CMake how to build the project. - The
tut1.ccfile contains the main function of the simulation and the most general structure of the simulation. - In the
model.hhfile we will define the numerical model, i.e. the set of differential equations that we want to solve. - The
parameters.jsonfile will contain the parameters for the simulation.
CMake setup
The CMakeLists.txt file is the first thing we need to set up. CMake is a powerful build system which automates complex setup of dependencies and structure of C++ programs. For a deeper understanding, please consult the documentation and see the DiFfRG build system setup in DiFfRG/cmake/setup_build_system.cmake.
In our case, we need to tell CMake about our new simulation and about the DiFfRG library. In practice, our CMakeLists.txt file looks as in the following:
In the above, we first request a minimum version of CMake and then declare our project.
Afterwards, we request CMake to find the DiFfRG package. We set the default install path here, which is /opt/DiFfRG, but depending on where you installed DiFfRG, you will have to change this.
Finally, we register an executable with CMake (called tut1) and use the setup_application function, which is a part of DiFfRG. It sets up all dependencies necessary for a DiFfRG-application, sets header paths and links it against the DiFfRG library.
With this set up, we can immediately create our build directory and test the build system. To that end, create a new directory and enter it,
Then, invoke cmake,
Here, we tell CMake to create an optimized "Release" build and propagate some further optimization flags to the compiler. The last argument to the cmake command, .., tells it where the source files can be found. In this case, it finds the CMakeLists.txt we just created in the parent directory and uses it to set up the build system.
At this point you could already invoke the build system by running
which will build the whole project using 8 cores. However, doing this will at this stage only lead to a linker error, as the tut1.cc file does not contain anything yet. In particular, the program lacks an entry point, i.e. a main function.
tut1.cc
Now we are ready to set up the main structure of the program in tut1.cc.
The includes in the first few rows are parts of the DiFfRG and we will use them in a moment to set up the simulation. Next, we include the model.hh file, where we will set up the actual system of equations to solve.
All relevant classes and functions are in the DiFfRG master namespace, so for convenience we just import all symbols from this namespace into our code by using namespace DiFfRG;.
The actual program logic starts with the entry point, the main function. The first thing to do in the main function is to read the parameter file parameters.json we created earlier:
The ConfigurationHelper class automatically chooses the correct parameter file; if the executable is invoked with the -p parameter, i.e. ./tut1 -p parameters.json, the argument of -p is chosen as the parameter file. Otherwise, ConfigurationHelper will try to open a file called parameters.json in the current working directory. The parameters have to be specified in the JSON format.
In order for this class to parse the flags and arguments passed from the command line, we need to construct the object with the argc, argv parameters, which hold this information. Afterwards, we load the entire parameter structure into the json variable, which has the same structure as the original json file.
After setting up the configuration, we choose the algorithms used in our simulation. To do so, we make some convenient type aliases with the chosen class types:
After making an alias for the numerical model, which we will specify in a moment, we choose a spatial (field space) dimension, which is here simply taken from the Model.
The Discretization gives a prescription to discretize the field space. Here, we choose the CG, i.e. Continuous Galerkin discretization and accordingly a fitting Assembler. The assembler, as the name implies, assembles the system of equations to be solved. In practice, the differential equation you specify in the Model is discretized by the Assembler using the Discretization and brought into the form of a time-dependent, nonlinear ODE (ordinary differential equation)
\[ F(\partial_t v_i, v_i) = 0\,, \]
where \(v_i\) are the components of the discretization. The Assembler is directly invoked by the TimeStepper which requests the construction of the above equation at every (RG) time step and evolves the system according to it. If you are unfamiliar with FEM methods, it would be recommended to at least understand the basics of it.
Next, we choose the SUNDIALS_IDA timestepper, which is the most performant choice for FEM setups with spontaneous symmetry breaking. IDA is a differential-algebraic solver, i.e. it solves both equations dependent on time, as above,
\[ F(\partial_t v_i, v_i) = 0\,, \]
but also stationary (secondary) equations without explicit dependence on time derivatives,
\[ G(v_i) = 0\,. \]
We will show the utility of this in a later tutorial. Note that the TimeStepper takes an additional argument, where we can choose the linear solver. The direct solver UMFPack is a simple but good choice, as it solves any linear system exactly. However, with large systems, e.g. in \(d\geq2\), GMRES or another iterative solver is usually much faster and thus preferred.
We now use the types we defined above to construct objects of all the classes described above.
With everything prepared, we are now ready to set up and run the equation system. To do so, we create an initial condition on the discretization (i.e. FEM space) we set up before,
and use it to run the time-stepper from RG-time 0 to the final RG-time, which we infer from the parameter file:
Note here, that RG-time is defined on the level of the code as positive, i.e.
\[ t_+ = \ln \Lambda / k \]
whereas the usual convention is
\[ t_- = \ln k / \Lambda \]
Furthermore, we recorded the time the simulation took using the dealii::Timer class.
We finish the program by printing a bit of information to the log file, in particular the performance and utilization of the Assembler.
model.hh
Now we need to specify the equation system to solve. For this example, we will just implement Burgers' equation, which is not fRG-related but a simple hydrodynamic equation given by
\[ \partial_t u(x,t) + \frac12 \partial_x u(x,t)^2 = 0 \]
First things first, we set a few things up before we get to the equation system:
Although not necessary in this case, the #pragma once compiler directive ensures that the header file is not included multiple times (similar to the use of include guards), which would lead to compiler errors. The DiFfRG/model/model.hh header contains basic predefines necessary for the setup of a numerical model. Again, we import all symbols from the DiFfRG namespace.
Afterwards, we create a struct to hold parameters we read once from the JSON variable. We call these a,b,c,d and will use them to construct the initial condition for the differential equation, i.e. \(u(x,0)\).
With this in place, let's set the equation system structure up:
Our system is very small, consisting only of a single equation. With large fRG simulations this can grow however extremely large, using momentum grids of variables and large vertex expansions. Thus, DiFfRG provides a mechanism of describing these systems and smartly indexing the numerical degrees of freedom. Here, we use a descriptor to tell the library how many and what kinds of functions we have. We use one function which lives on a FEM discretization and thus use the FEFunctionDescriptor with one Scalar which we call "u" as in the above equation.
If we had two variables, e.g. another one called \(v\), we would declare this as FEFunctionDescriptor<Scalar<"u">, Scalar<"v">>;. We will treat the declaration of larger systems in a later tutorial in-detail.
The ComponentDescriptor type packs the FEFunctionDescriptor together with other parts of the equation system; here, we only have a FE function and thus do not provide more template parameters. Creating a compile-time constant (constexpr) object of the descriptor is useful for easier access to the indices, which we will use below.
Now we define the actual numerical model. We need to derive the class from def::AbstractModel, which defines all necessary methods so our numerical model can be used by the Assembler. Here, we also communicate the structure of our equation system through the Components type we declared earlier. Furthermore, we derive from a few more classes:
def::fRGprovides a method where \(t\) is automatically communicated to the model and the RG-scale \(k\) is computed from it. Furthermore, it defines the variablestandkwhich can be directly used in the code.def::FlowBoundariesis necessary to close the FEM system, as the boundary conditions at \(x=0\) and \(x = x_\textrm{max}\) need to be specified.def::FlowBoundariessets up inflow/outflow boundary conditions.def::ADcreates methods for the automatic evaluation of jacobians of our system, i.e.\[ \frac{\partial (\partial_t u - \frac12 \partial_x u^2)}{\partial u}\,\qquad\text{and}\qquad \frac{\partial (\partial_t u - \frac12 \partial_x u^2)}{\partial (\partial_t u)}\,. \]
This is achieved by the use of automatic differentiation, using the autodiff library. This is also the reason why we must template and not use explicit types in the methods of the numerical model, as will be seen below.
Here, we have only written the constructor, communicated \(\Lambda\) to the def::fRG base class and created a Parameters object.
The initial_condition method is invoked by the initial_condition.interpolate(model) call from above. We use the parameters a,b,c,d to set up a polynomial initial condition. Here, we also use the indexing provided by the FEFunctionDescriptor class, to retrieve the position of "u" within the array of FE functions. In this case, it is of course trivial, as idxf("u") always evaluates to 0. Note however, that the lookup is done statically at compile time and is thus much faster than dynamic lookup, such as with a std::map.
The flux method implements the actual equation. If we don't specify anything else, the Assembler will construct the equation
\[ \partial_t u + \partial_x (F_i(u)) = 0 \]
so that our choice of \(F_i(u) = - \frac12 u^2\) exactly implements Burgers' equation.
parameters.json
The parameter file contains usually user-defined quantities in a "physical" subsection and further paramters for the backend:
These sections are just the parameters we also use in the numerical model, i.e. user-space parmeters.
The discretization section configures the FEM setup of our simulation:
fe_ordersets the polynomial order of the local function space on the finite elementsthreadssets the number of CPU threads used for assembly. Note, that other multithreading parts ofDiFfRG, such as momentum integration, automatically use all available CPU threads and ignore this parameter.batch_sizethe assembly threads get batches ofbatch_sizewhich they sequentially process. Playing around withthreadsandbatch_sizemay give a small performance boost, but keepingthreadsaround the number of physical cores andbatch_sizearound 32-64 should be sufficient for almost optimal performance.overintegrationcan be used to increase the order of the quadratures used in assembly when constructing the weak form of the PDE. It is seldom necessary to increase beyond 0.output_subdivisionsgives the precision with which the grids in the output data are written. This goes exponentially, so don't choose it too high.EoM_abs_tolsets the absolute precision within which a specified equation of motion is solved. DiFfRG can be instructed to solve the EoM at every timestep and perform additional computations at this point.EoM_max_itersets the number of bisections used in determining the position of the EoM.x_gridthis works in a python-like slice syntax and sets the used in aRectangularMesh. The parameter also supports locally different cell sizes: "0:1e-4:1e-2, 1e-2:1e-3:1" creates 100 cells between 0 and 1e-2, and 100 cells between 1e-2 and 1.y_gridandz_gridwork identically, but are only used in 2D / 3D simulations.refinecan be used to quickly increase the cell count by \(2^\textrm{refine}\).The adaptivity section sets parameters for adaptive refinement of the discretization mesh. As we have not set this up in the numerical model, we will postpone the explanation of this section to a later tutorial."adaptivity": {"start_adapt_at": 0E0,"adapt_dt": 1E-1,"level": 0,"refine_percent": 1E-1,"coarsen_percent": 5E-2}},The timestepping section controls the behavior of the"timestepping": {"final_time": 10.0,"output_dt": 1E-1,"explicit": {"dt": 1E-4,"minimal_dt": 1E-6,"maximal_dt": 1E-1,"abs_tol": 0.0,"rel_tol": 1E-3},"implicit": {"dt": 1E-4,"minimal_dt": 1E-6,"maximal_dt": 1E-1,"abs_tol": 1E-13,"rel_tol": 1E-7}},Timestepperclass:final_timesets the end-time of the simulation - this is actually not a required parameter, but one we use in our call totimestepper.run(...)output_dtsets the intervals within which we write the solution to disk. Note, that the timesteppers do not necessarily step exactly ontooutput_dtintervals, but interpolate the solution to the output times.- The
explicitandimplicitsections control the behavior of explicit and implicit timesteppers respectively. The disctinction exists, because some timesteppers actually run part of the equation system explicitly and another implicitly. Usually, the FEM part is always implicit and further components of the expansion can be chosen to evolve explicitly.dtis the initial timestep for adaptive timesteppers and the fixed timestep for non-adaptive ones.minimal_dtandmaximal_dtset the bounds for the timestep size - if an adpative stepper goes above or below, the step is adjusted to the bound, if the timestepper gets stuck belowmminimal_dt, time evolution is aborted.abs_tolandrel_tolset absolute and relative error tolerance for the timestepping. While the FE parts of the system usually require a higher precision (< 1e-7 relative precision), especially in the presence of spontaneaous symmetry breaking, explicit tolerances can be chosen much more freely. For more information about timesteppers and their relationship with spontaneous symmetry breaking in fRG, see also this paper.
The output section defines:
verbositysets how much information is being written to console while running. If at 0, no information is written at all, whereas at 1 the system gives updates at every timestep.foldersets the base folder where data is stored. This is useful to not clutter your current directory ("./") with output, or if you run a large amount of simulations.namesets the beginning of all filenames of the output, i.e. in this case all files created by the simulation start with "output".
Running it
To run the simulation, first build it,
and then directly run it through
We made use here of a mechanism to override the parameters of a simulation. The flag -p selects the parameter file to be used. By default it looks for a parameter.json in the current directory. One can use the flags -si to set an integer, -ss to set a string and -sd to set a double (floating-point) value, followed by the full path to the parameter and its value after an equality symbol.
After the simulation has run, which should take (depending on your computer) a few seconds, a few files have been created in the current folder, which you can look up by
output.logcontains logging information:$ cat output.log[2024/10/11] [23:01:31] [DiFfRG Application started][2024/10/11] [23:01:31] [FEM: Number of active cells: 100][2024/10/11] [23:01:31] [FEM: Number of degrees of freedom: 301][2024/10/11] [23:01:31] [FEM: Using 8 threads for assembly.][2024/10/11] [23:01:32] [CG Assembler:Reinit: 4.36692ms (1)Residual: 0.184398ms (3557)Jacobian: 1.24521ms (17)][2024/10/11] [23:01:32] [Simulation finished after 1s]output.log.jsoncontains a copy of the JSON parameters used.output.pvdcontains links to the FEM output data of the simulation and can be now visualized inParaview.
After invoking $ paraview output.pvd, we open a new tab and choose line chart view:
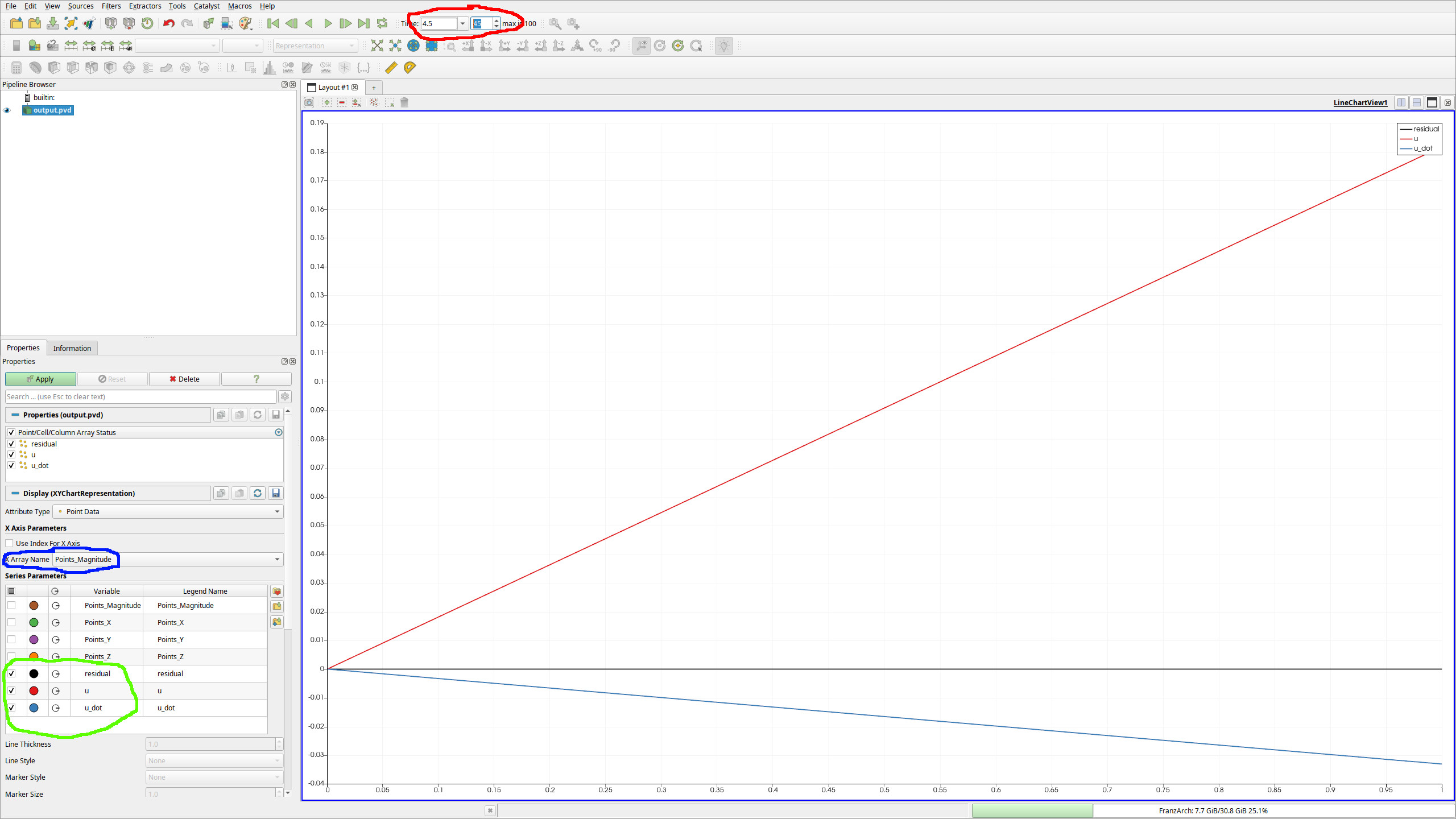
- At the red marked field, one can change the currently displayed time of the FEM function.
- The field marked in blue defines what the x-axis is. The default sets this to indices, but this does not work well in general and especially for irregularily spaced grids. Therefore, one would usually set this to
Points_Magnitudefor a 1D FEM simulation. - In the green marked part you see that we have the FE functions
u,residualandu_dotwhich we can look at.uof course is just the solution to Burgers' equation,u_dotis its time derivative, andresidualis the local error of the equation.
Feel free to play around also with the parameters a,b,c,d and other parts of the parameters in order to get acquainted with the basics of the framework.